 Multiple Choice Questions
Multiple Choice QuestionsWhich statement is correct regarding ABC transporters?
Consist of the transmembrane domain as well as the nucleotide-binding domain
All are P-glycoproteins
Present in only eukaryotes
Makes membrane porous
In prokaryotes during replication, the lagging strand is synthesized in a series of short fragments known as Okazaki fragments, consequently requiring many primers. The RNA primers of Okazaki fragments, consequently requiring many primers. The RNA primers of Okazaki fragments are subsequently degradaed by DNA Polymerase I and the gap are filled. How DNA Polymerase I fills the gap once the primer have been removed from lagging strand?
DNA Polymerase I has its own primer
DNA Polymerase I do not require primer
DNA from leading strand serves as primer
Ends of existing Okazaki fragments on lagging stand serves as primer
Substrate for DNA synthesis is?
Nucleotide tri phosphate
Nucleoside tri phosphate
Nucleoside pyrophosphate
Ribonucleotide tri phosphate
The 5’ Cap of RNA is required for the
stability of RNA only
Stability and transport of RNA
Transport of RNA only
Methylation RNA
B.
Stability and transport of RNA
5'cap protects mRNA against ribonuclease degradation, fuction in the initiation of protein synthesis and transport of RNA.
Presence of circular mRNA for a specific protein in an eukaryotic cell reflects a rapid rate of synthesis of that protein. Following mechanism are suggested:
a. eIF‐4G and PABP promote this process through 5’‐3’ interaction of mRNA
b. Ribosome are less active in recognizing circular mRNA
c. PABP and eIF‐4A promote this process
d. Ribosome can reinitiate translation without being disassembled
Which of the following is correct?
A and D
B and D
A and C
B and C
siRNA and miRNa are used for achieving gene silencing. Although, major steps are similar there are distinct differences in the key players of the two processing pathways. Following statements related to some characteristic features of gene silencing
A. Both siRNAs and miRNAs are processed by cytoplasmic endonuclease dicer
B. Drosha is needed for processing miRNa and precursor siRNa
C. Both siRNA and miRNA show association with Argonaute protein
D. Both the processing pathways involve RISe complex
Which of the following combinations is NOT correct?
A and C
C and D
A and B
D and A
In eukaryotic chromatin, 30 nm fibre (solenoid) can open up to give rise to two kinds of chromatin. In one type (A), the promoter of a gene within the open chromatin is occupied by a nucleosome whereas in the other (B), the promoter is occupied by histone H1. The following possibilities are suggested
A. The gene in (A) is repressed
B. The gene in (B) is repressed
C. The gene in (A ) is active
D. The gene in (B) is active
Which of the following sets is correct?
A and B
A and B
B and D
C and D
Genetic studies demonstrated that TBP mutant cell extracts are deficient in transcription of genes from all three promoters viz. class I, II, and III. Following statements describe characteristic features of TBP.
A. TBP is considered as a universal basal transcription factor
B. TBP is not required for transcription of archaeal genes
C. TBP is involved in recognizing TATA box
D. TBP operates at all promoters regardless of their TATA content
Which of the following combinations is NOT correct?
A and D
C and D
B and D
A and C
Based on the structural regions of a nuclear receptors shown in the diagrams, the following predicts were made
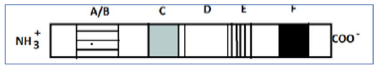
A. Region F is responsible for binding to ligands and conditions two zinc finger‐like binding motifs.
B. Receptors with A/B domains generally associate with chaperons and do not bind to DNA.
C. Region E indicate that receptors associate with chaperones which protect the nuclear hormone receptors.
D. Region C contains the P‐bod and the D‐box required for dimerization of the receptor and creates contact with DNA phosphate backbone.
Which one of the following is true?
A and B
B and C
B and D
C and D
