 Multiple Choice Questions
Multiple Choice QuestionsReverse transcriptase has both ribonuclease and polymerase activities. Ribonuclease activity is required for?
The synthesis of new RNA strand.
The degradation of RNA strand.
The synthesis of new DNA strand
The degradation of DNA strand
Amino acid selenocysteine (Sec) is incorporated into polypeptide chain during translation by
Charging of Sec into tRNAser followed by incorporation through serine codon.
Charging of serine into tRNAser followed by modification of serine into selenocysteine and then incorporation through serine codon.
Charging of sec into tRNAsec and then incorporation through selenocysteine codon.
Charging of serine into tRNAser, modification of serine into selenocysteine and then incorporation through a specially placed stop codon.
D.
Charging of serine into tRNAser, modification of serine into selenocysteine and then incorporation through a specially placed stop codon.
Selenocysteine (Ser) isincorporated into polypeptide chain during translation by charging of serine into tRNA through a specially placed stop codon.
α-amanitin inhibits
Only RNA polymerase I
Only RNA polymerase II
Only RNA polymerase III
All RNA Polymerases
While replicating DNA, the rate of mis-incorporation by DNA Polymerase is 1 in 103 nucleotides. However, the actual error rate in the replicated DNA is 1 in 109 nucleotides incorporated. This is achieved mainly due to
Spontaneous excision of mis-incorporated nucleotides.
3’ → 5’ proof reading activity of DNA polymerase.
Termination of DNA polymerase at mis-incorporated sites.
5’ → 3’ proof reading activity.
A single strand nick in the parental DNA helix ahead of a replication fork causes the replication fork to break. Recovery from this calamity requires
DNA ligase
DNA primase
Site-specific recognition
Homologous recombination
Denaturation profiles of DNA are shown below.
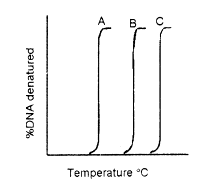
The differences in the profiles arise because
The DNA is singe stranded but of different sizes.
A + T content of A > B> C and the DNA are from complex genomes.
G + C content of C > B> A in DNA of comparable sizes isolated from simple genomes.
G + C content is identical but A + T content in A > B > C in DNA of comparable sizes isolated from simple genomes.
In human, protein coding genes are mainly organized as “exons” and “introns”. There are intergenic regions that transcribe into various types of non-coding RNA (not translating into protein). Some introns may harbor also transcription units, which are
Always other protein coding genes
Protein coding gene and RNA coding genes
Always RNA coding genes
Pseudo genes
Pre-mRNAs are rapidly bound by snRNPs which carry out dual steps of RNA splicing, that removes the intron and joins the upstream and downstream exons.
The following statements describe some facts related to this event.
A. Almost all introns begin with GU and end with AG sequences and hence all the GU or AG sequences are spliced out of RNA.
B. U2 RNA recognizes important sequences at the 3’receptor end of the intron.
C. The spliceosome uses ATP to carry out accurate removal of intron.
D. An unusual linkage with 2’OH group of guanosine within the intron form a ‘Lariat’ structure.
Which of the following combinations is correct?
A and B
B and C
C and D
D and A
For continuation of protein synthesis in bacteria, ribosomes need to be released from the mRNA as well as to dissociate into subunits. These processes do not occur spontaneously. They need the following possible conditions:
A. RRF and EF-G aid in this process
B. An intrinsic activity of ribosomes and an uncharged tRNA are required
C. IF-1 promotes dissociation of ribosomes
D. IF-3 and IF-1 promote dissociation of ribosomes
Which of the following sets is correct?
A and D
A and B
A and C
B and D
Insulin and other growth factors stimulate a pathway involving a protein kinase mTOR, which in its turn augments protein synthesis. mTOR essentially modifies protein (s) which in their unmodified form act as inhibitors of protein synthesis. The following proteins are possible candidates:
A. eEF-1 B. eIF-4E-BP1 C. eIF-4E D. PHAS-1
Which of the following sets is correct?
A and B
B and D
A and C
B and C
